|
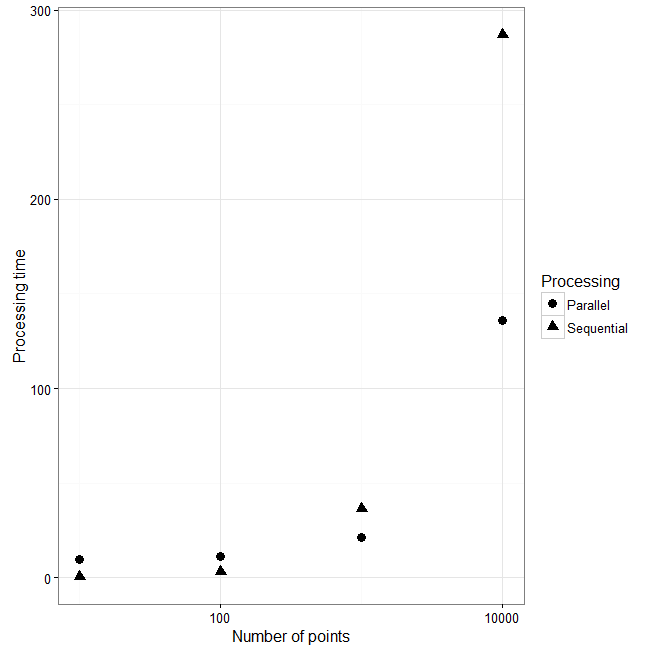
In the following experiment, I first created a spatially autocorrelated landscape that might represent a Digital Elevation Model (DEM). This landscape consist of 250,000 pixels that range from 500 to 1000 units. You should note that this is a pretty small landscape compared to what you would have from real data. Most DEM I have worked with have > 1 million pixels. Consequently, things would take way longer. Just to check how long it takes to run through this simple landscape and collect data for an n set of zones, I sequentially increase the number of points and extract the data from a 100 unit buffer. I also calculate some statistics on this data using the summary() and sd() function. In the graph on the right, you'll see that as I increase the number of points, the processing time increases ~ 10 seconds to 300 (Triangles). If I do this again using my parallel function the amount of processing time is about the same when performing zonal statistics on 10 and 100 spatial points (Circles). However, the parallel functions is much quicker for 1000 and 10,000 spatial point buffers. It actually cuts the time in haft for 10,000 point buffers (From 290 seconds to ~140).
I wrote earlier that the processing time should be 4x quicker compared to sequential processing. However, my experiment only decreased processing time by 2. The reason is that we haven't really reached the point where this type of parallel processing is performing at its best. |
|
If you ever had to compute zonal statistics for several polygons or point buffers (> 100) you may have noticed that it takes a very long time to perform this in R and in a GIS such as ArcGIS. And as the number of zones and the number of pixels representing your landscape increases, so does computing time (sometimes exponentially). One way to make this process quicker is to create your own parallel version of zonal statistics in R. You can thread the number of sequential operations to the number of CPUs you have in your computer. For instance, if I had 1000 zones and 4 CPUs, I would split these zones into 4 lists of 250 zones and run them on 4 of my CPUs. In additional, you can run batches of these points to reduce the amount of memory. For example I would split my 1000 points into 4 sets of 250 points and I would also take these 250 points from each set and create 25 batches of 10 points. if you have enough memory and everything works out well, you can increase the processing speed up to 4x faster or even 8 if you have 8 CPUs. For this type of work I use the snowfall package in R. I first create a function that I can run sequentially using the lapply() function. Then I try it with the snowfall sfLapply() function. There aren't any major modification you need to do. In most cases you simply need to start up your cluster by doing sfInit(parallel = T, cpus = 4), replace lapply with sfLapply and run the function and remember to do sfStop() to turn your cluster off. This is for a windows machine. For those running on a mac or linux OS you can use the mclapply function in the parallel package. It is much easier to work with compared to snowfall. You simply have to change the lapply() function to mclapply() and add in the mc.cores = 4 parameter to split everything on 4 threads. Also you might want to note these little differences in each function lapply(X, FUN, ...), sfLapply(x, fun, ...) and mclapply(X, FUN, ...).
Eventually, I would expect the difference between parallel and sequential processing time to continuously increase as we add more points. Note that I didn't test it on 100,000 points because my laptop ran out of memory. This is one issue with running code in parallel, it is very memory intensive and you need to make your code memory efficient, since your running the code n times at once. For parallel processing in this example, the landscape is replicated in my virtual memory the same amount of time as the number of CPUs. Consequently, my parallel code takes up 4x more memory then if we ran it sequentially. To decrease the amount of virtual memory, you can modify this by further threading the points within each CPU so it runs in batches of k number of points. The code I wrote splits the points up into number of points / number of CPUs. This means if I have 10,000 points, it creates 4 batches of points containing 2,500 points. You can change this by modifying the bsize parameter within the code. If you are interested in working with other types of zones other than point buffers, you can easily adapt this code to run on polygons by simply replacing spatial point objects with spatial polygons. Feel free to email me about transforming this code to work on polygons.
Comments are closed.
|
Archives
July 2016
Robby R. MarrotteThis section contains bits of code I use in my research and some observations I've made in the field. Categories
All
|
 RSS Feed
RSS Feed
